Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Ishida, Hisashi; Matsumoto, Atsushi; Tsutsumi, Yu*; Yura, Kei
Proceedings of 16th International Microscopy Congress (IMC 2006), P. 242, 2006/09
Supra-biomolecules, which contain numerous proteins and nucleic acids, function when the constituent molecules are assembled. Therefore, it is important to determine not only the 3D structures of the constituent molecules but also the 3D structure of the supra-biomolecule. Although X-ray crystallography can determine the atomic coordinates of biomolecules, it has difficulty in handling supra-biomolecules, because crystals of huge molecules cannot be made with ease. Single particle analysis using an electron microscope (EM) has been used to observe the structure of supra-biomolecules, but the resolution of the EM image has only achieve to the atomic level in a limited situation. Therefore, several attempts have been carried out to determine the 3D structure of supra-biomolecules in atomic resolution by fitting the constituent molecules, determined by X-ray crystallography, into an EM density map. In those attempts, each constituent molecule is usually fit into the EM density map manually, and the constituent molecules may have atomic collisions at their interfaces.
Ishida, Hisashi; Matsumoto, Atsushi; Tsutsumi, Yu*; Yura, Kei
no journal, ,
We are developing an EM density-fitting refinement method to improve the modeled 3D structure of supra-biomolecules by alleviating the steric stress of atoms while retaining the condition that the constituent molecules fit in the EM density map. The method was applied to ribosome. We used an atomic structure of Thermus thermophilus 70S ribosome which was determined by X-ray crystallography (PDB code: 1YL3 and 1YL4). The target EM data from Escherichia coli 70S ribosome, which were observed under various reaction conditions from the initial to the final stage of the translation of messenger RNA, were retrieved from the electron microscopy database at the European Bioinformatics Institute. In our method, the X-ray structure of ribosome was fit into the EM maps as a rigid-body by matching the three inertial axes of the X-ray structure with those of each EM map first. Then, the atomic coordinates were transformed along the directions of low frequency modes calculated by NMA until the best fit was achieved. Finally, MD simulations in which ribosomal RNAs were treated as flexible molecules and the other molecules were treated as rigid-body molecules were performed to produce a refined 3D structure of ribosome without atomic collisions. The MD simulations also optimized the match between the modeled 3D structures and the EM maps. It was found that the NMA simulations could match the 3D structure and the EM maps in the range of 59-78%, while the MD simulation matched them in the range of 56-82%. In the MD simulation, the center of masses and orientations of constituent molecules of ribosome in the X-ray structure and the refined structure were compared, and the amplitudes of the differences of the centers of masses and the orientations was about 2 Å and about 4º respectively on average. It is considered that the structural changes indicate the functional movements in ribosome under various reaction conditions.
 coaxial nanotubes
coaxial nanotubesTaguchi, Tomitsugu; Igawa, Naoki; Yamamoto, Hiroyuki; Shamoto, Shinichi
no journal, ,
no abstracts in English
Terauchi, Masami*; Koike, Masato; Fukushima, Kurio*; Kimura, Jun*
no journal, ,
no abstracts in English
 irradiation
irradiationHojo, Tomohiro*; Yasuhara, Akira*; Yamamoto, Hiroyuki; Aihara, Jun; Furuno, Shigemi*; Sawa, Kazuhiro; Sakuma, Takashi*; Hojo, Kiichi
no journal, ,
Yttria stabilized zirconia (YSZ) is a candidate material as optical and insulating materials in nuclear reactors. The investigation of defect formation under the ion irradiation is therefore required to assess YSZ resistance for radiation damage. Ion irradiation and nanostructural observations were simultaneously performed by transmission electron microscope (TEM) equipped with ion accelerator and X-ray spectrometer (EDS). YSZ was irradiated with 30 keV Ne ions with a flux of 3
ions with a flux of 3 10
10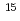 ions/cm
ions/cm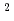 at 923 K. It is clearly observed that the dislocation loops are formed along (110) crystal plane. The measurements across the loops by EDS show that the composition of oxygen on loops decreases and zirconium increases compared with the matrix. It is also shown that the intensity of Ne increases at the loop. From the previous studies, it was observed that bubbles were formed preferentially on loop planes. It is implied that the irradiated Ne tends to exist in the loops as bubbles.
at 923 K. It is clearly observed that the dislocation loops are formed along (110) crystal plane. The measurements across the loops by EDS show that the composition of oxygen on loops decreases and zirconium increases compared with the matrix. It is also shown that the intensity of Ne increases at the loop. From the previous studies, it was observed that bubbles were formed preferentially on loop planes. It is implied that the irradiated Ne tends to exist in the loops as bubbles.